Use Google's Earth Engine Python API in RStudio
This blogpost is about RStudio and the reticulate package! Reticulate includes a Python engine for R Markdown that enables easy interoperability between Python and R chunks. This enables us to bring the power of Earth Engine to RStudio.
Tutorial: Deriving simple tree phenology data from Sentinel2 with Earth Engine and plotting the data in R.
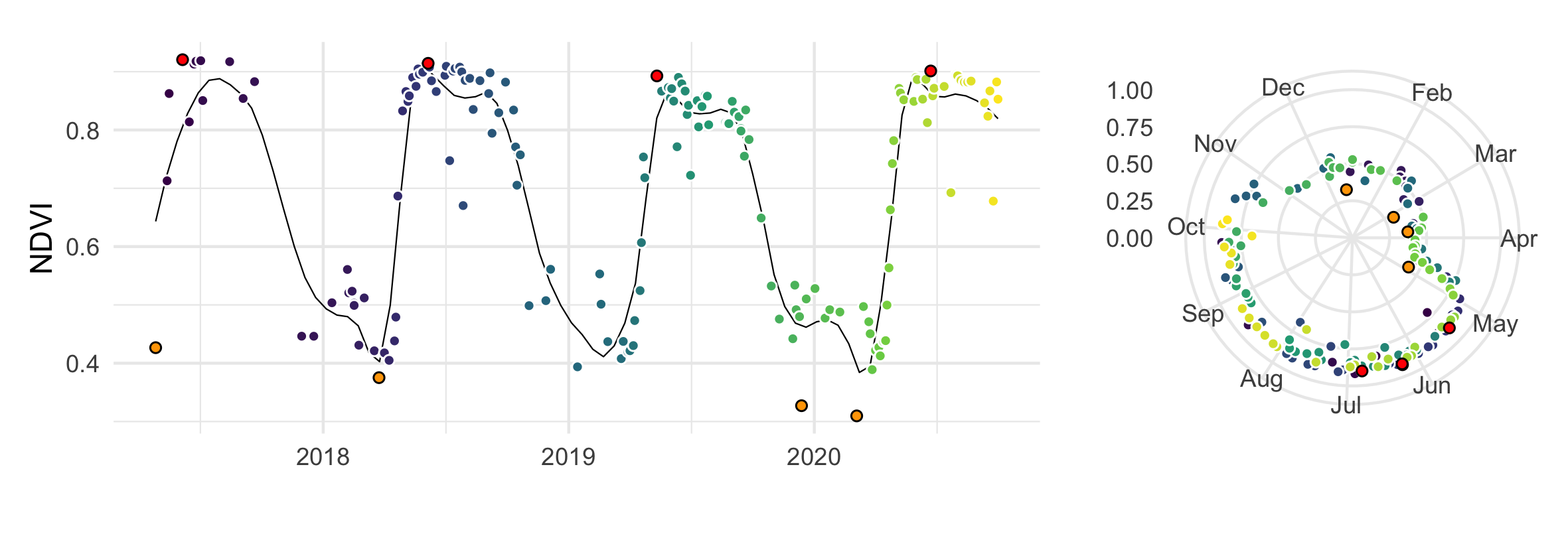 Sentinel-2 derived NDVI values of a beech forest plotted over four years with annual min/max values highlighted.
Sentinel-2 derived NDVI values of a beech forest plotted over four years with annual min/max values highlighted.
Life between Use GoogleEarthEngine and in RStudio
Life between #GoogleEarthEngine and #R pic.twitter.com/ZbI33X21i3
— Hammad Gilani (@gilanius) October 14, 2020
With reticulate we can use:
- the Google Earth Engine Python API to extract values of satellite data (in Python chunks),
dplyr&ggplotpackages to manipulate the extracted data and to create e.g. a phenology chart (in R chunks).
My Setup
- Mac - macOS High Sierra 10.13.6
- R - version 3.6.3
- RStudio - version 1.3.1093
- Conda - version 4.7.12
Create new conda environment with earthengine-api
The RStudio Terminal provides access to the system shell from within the RStudio IDE. Open your Terminal tab and write:
### Terminal
# Create a conda environment
conda create --name gee-demo
# Activate the environment
conda activate gee-demo
# Install the Earth Engine Python API
conda install -c conda-forge earthengine-api
# Authenticate your access with the command line tool
earthengine authenticate
conda install pandas
conda install numpyCreate new R Markdown file
In RStudio, create a new R Markdown file (File => New File => R Markdown …) and install the reticulate package and apply the use_condaenv() function to point to the created ‘gee-demo’ conda environment.
### R
if(!require("reticulate")) install.packages("reticulate")
library(reticulate)
use_condaenv("gee-demo", conda = "auto",required = TRUE)The Earth Engine Python API is available as a Python package that can be installed locally. The Python API package is called ee. It must be imported and initialized for each new Python session and script.
### Python
import ee
ee.Initialize() # Initialize the API
print(ee.__version__)
## 0.1.238At this point we have set up our working environment and can get started. The ultimate goal is to use this working environment and to visualize the phenology of a beech forest without leaving RStudio.
Generate NDVI data for a german beech forest
With the help of the opentrees.org project I found a beech forest near Potsdam in East Germany. Opentrees.org is the world’s largest database of municipal street and park trees, produced by harvesting open data from dozens of different sources.
### Python
geometry = ee.Geometry.Polygon( # beech forest area
[[[13.109782839513642, 52.42163736335469],
[13.109782839513642, 52.420752422262055],
[13.111671114660126, 52.420752422262055],
[13.111671114660126, 52.42163736335469]]]);Create a function to mask clouds using the Sentinel-2 QA band
### Python
def maskclouds(image):
qa = image.select("QA60")
cloudBitMask = 1 << 10
cirrusBitMask = 1 << 11
mask = qa.bitwiseAnd(cloudBitMask).eq(0) \
.And(qa.bitwiseAnd(cirrusBitMask).eq(0))
return image.updateMask(mask).divide(10000) \
.set('date', image.date().format()) # adding recording dateCreate a function to add Normalized Difference Vegetation Index (NDVI) band
### Python
def ndvi(image):
ndvi = image.normalizedDifference(["B8","B4"]).rename("ndvi")
return image.addBands([ndvi])Create two ImageCollections with Sentinel2 Level-2A and cloud probability data
### Python
start_date = ee.Date('2017-04-01')
end_date = ee.Date('2020-12-31')
s2Sr = ee.ImageCollection("COPERNICUS/S2_SR") \
.filterBounds(geometry) \
.filterDate(start_date, end_date) \
.filter(ee.Filter.eq('MGRS_TILE', '33UUU')) \
.map(maskclouds) \
.map(ndvi)
s2c = ee.ImageCollection('COPERNICUS/S2_CLOUD_PROBABILITY') \
.filterBounds(geometry) \
.filterDate(start_date, end_date);Create a function to Join two collections on their ‘system:index’ property.
### Python
# The propertyName parameter is the name of the property
# that references the joined image.
def indexJoin(collectionA, collectionB, propertyName):
joined = ee.ImageCollection(ee.Join.saveFirst(propertyName).apply(
primary = collectionA,
secondary = collectionB,
condition = ee.Filter.equals(
leftField = 'system:index',
rightField = 'system:index'
)
))
# Merge the bands of the joined image.
return joined.map(lambda image: image.addBands(ee.Image(image.get(propertyName)).rename("cld")))Create a function to get a) median value and b) the recording date for each image
### Python
def calculateMedian(image):
median = image.reduceRegion(reducer = ee.Reducer.median(),
geometry = geometry,
scale = 10,
maxPixels = 1e9)
# set the recording date and median values as a property of the image
return image.set('date', image.get('date')).set('median', median)Apply the Join function and median values
### Python
withCloudProbability = indexJoin(s2Sr, s2c, 'cloud_probability');
# select only the 'ndvi' and 'cloud probability' values.
withMedian = withCloudProbability.select(["ndvi","cld"]).map(calculateMedian)
# reduce the images properties to a list of lists
values = withMedian.reduceColumns(ee.Reducer.toList(2), ['date', 'median']).values().get(0)
# Type casts the result into a List
eeList = ee.List(values)
# convert list of lists to a dictionaty
medians = ee.Dictionary(eeList.flatten())Call getInfo() on Earth Engine objects to get the object from the server to the client
### Python
median_values = medians.getInfo(); # ee.Dictionary => Python dictionary
type(median_values)
## <class 'dict'>Python dictionary to pandas DataFrame
### Python
import pandas
pd_df = pandas.DataFrame.from_dict(median_values, orient='index')STOP - lets see what we have got - STOP
### Python
pd_df
## cld ndvi
## 2017-04-24T10:11:20 84.0 NaN
## 2017-04-27T10:20:25 9.0 0.426655
## 2017-05-04T10:13:49 100.0 NaN
## 2017-05-14T10:11:25 16.0 0.712856
## 2017-05-17T10:23:52 1.0 0.862517
## ... ... ...
## 2020-09-30T10:16:16 6.0 0.852820
## 2020-10-03T10:26:13 67.0 NaN
## 2020-10-08T10:26:16 98.0 NaN
## 2020-10-10T10:16:16 99.0 NaN
## 2020-10-13T10:26:14 100.0 NaN
##
## [457 rows x 2 columns]In total we have NDVI and cloud probability data (from cloud free [0 %] to cloudy [100 %]) of 457 Sentinel-2 images in our DataFrame. Some rows have NaN values in the NDVI column. NaN values occur if clouds were recognized in the QA60 band and masked out.
Calling Python from R
The pd_df panda DataFrame, created within the Python chunk above, is available to R using the py object from the reticulate package.
### R
df <- py$pd_df # convert pandas DataFrame to R DataFrame
head(df)
## cld ndvi
## 2017-04-24T10:11:20 84 NaN
## 2017-04-27T10:20:25 9 0.4266549
## 2017-05-04T10:13:49 100 NaN
## 2017-05-14T10:11:25 16 0.7128555
## 2017-05-17T10:23:52 1 0.8625174
## 2017-05-24T10:13:53 100 NaNPrepare data for plotting
For the work with R we use the following packages:
### R
list.of.packages <- c("stringr", "dplyr", "tidyr", "ggplot2", "patchwork")
new.packages <- list.of.packages[!(list.of.packages %in% installed.packages()[, "Package"])]
if (length(new.packages)) install.packages(new.packages)
library(stringr)
library(dplyr)
library(tidyr)
library(ggplot2)
library(patchwork)In the next code chunk we manipulate the data to make our life easier with the plotting. We extract the recording date from the rownames and additionally create year and doy (day of the year) values. We also delete all lines with NaN NDVI values and exclude images with cloud probability values greater than 40.
### R
xx <- df %>%
mutate(row_names = rownames(df)) %>%
mutate(datetime = stringr::str_split(row_names, "T", simplify = TRUE)[,1]) %>%
select(-row_names) %>%
drop_na(ndvi) %>%
mutate(date = as.Date(datetime, format="%Y-%m-%d")) %>%
filter(cld < 40) %>%
mutate(yday = lubridate::yday(date)) %>%
mutate(year = lubridate::year(date)) %>%
mutate(id = 1:n())
str(xx)
## 'data.frame': 164 obs. of 7 variables:
## $ cld : num 9 16 1 2 21 22 11 7 16 4 ...
## $ ndvi : num 0.427 0.713 0.863 0.921 0.814 ...
## $ datetime: chr "2017-04-27" "2017-05-14" "2017-05-17" "2017-06-06" ...
## $ date : Date, format: "2017-04-27" "2017-05-14" ...
## $ yday : num 117 134 137 157 167 174 177 184 187 227 ...
## $ year : num 2017 2017 2017 2017 2017 ...
## $ id : int 1 2 3 4 5 6 7 8 9 10 ...
dim(xx)
## [1] 164 7After filtering all NaN values and exclude cloud probabilities higher than 40 % we have 164 rows (median NDVI values) left to work with.
Plotting the data
### R
max <- xx %>%
group_by(year) %>%
filter(ndvi == max(ndvi))
min <- xx %>%
group_by(year) %>%
filter(ndvi == min(ndvi))
plot_title <- expression(paste("Phenology of a beech forest ", italic("(Fagus Sylvatica)"), " near Potsdam - Germany"))
plt1 <- ggplot(data = xx, aes(x = date, y = ndvi, fill = id)) +
geom_smooth(colour = "black", size= 0.25,se = FALSE, span = 0.15)+
geom_point(colour = "white",pch=21) +
geom_point(data = max, aes(x = date, y = ndvi),fill = "red", pch = 21) +
geom_point(data = min, aes(x = date, y = ndvi),fill = "orange", pch = 21) +
ggtitle(plot_title) +
xlab("") + ylab("Normalized Vegetation Index (NDVI)") +
scale_fill_viridis_c() + theme_minimal() + theme(legend.position = "none")
plt1### R
plt2 <- ggplot(xx, aes(x=yday, y=ndvi, fill = id))+
coord_polar(theta="x", start=0,direction=1) +
geom_point(colour = "white", pch = 21) +
geom_point(data = max, aes(x = yday, y = ndvi),fill = "red", pch = 21) +
geom_point(data = min, aes(x = yday, y = ndvi),fill = "orange", pch = 21) +
scale_y_continuous(limits = c(0, 1)) +
xlab("") + ylab("") +
labs(caption = "Data - Sentinel2 | Processed - EarthEngine Python API") +
scale_x_continuous(breaks = 30*0:11, minor_breaks = NULL, labels = month.abb) +
scale_fill_viridis_c() + theme_minimal() + theme(legend.position = "none")
plt2 ### R
plt1 + plt2 + plot_layout(widths = c(2.5, 1), heights = unit(c(5), c('cm', 'null')))Other examples
Session Info
### R
sessionInfo()
## R version 3.6.3 (2020-02-29)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS High Sierra 10.13.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] patchwork_1.0.0 ggplot2_3.3.2 tidyr_1.1.0 dplyr_1.0.0
## [5] stringr_1.4.0 reticulate_1.16
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.5 pillar_1.4.6 compiler_3.6.3 tools_3.6.3
## [5] digest_0.6.25 nlme_3.1-144 jsonlite_1.7.1 lubridate_1.7.9
## [9] evaluate_0.14 lifecycle_0.2.0 tibble_3.0.4 gtable_0.3.0
## [13] lattice_0.20-38 viridisLite_0.3.0 mgcv_1.8-31 pkgconfig_2.0.3
## [17] rlang_0.4.8 Matrix_1.2-18 yaml_2.2.1 xfun_0.18
## [21] withr_2.3.0 knitr_1.30 generics_0.0.2 vctrs_0.3.4
## [25] grid_3.6.3 tidyselect_1.1.0 glue_1.4.2 R6_2.4.1
## [29] rmarkdown_2.3 farver_2.0.3 purrr_0.3.4 magrittr_1.5
## [33] splines_3.6.3 scales_1.1.1 ellipsis_0.3.1 htmltools_0.5.0
## [37] colorspace_1.4-1 labeling_0.3 stringi_1.5.3 munsell_0.5.0
## [41] crayon_1.3.4Bottom line
This blogpost showed you how to use Earth Engine Python API in RStudio. If you have any questions, suggestions or spotted a mistake, please use the comment function at the bottom of this page.
You can cite this blogpost using: Gärtner, Philipp (2020/10/16) “Use Google’s Earth Engine Python API in RStudio”, https://philippgaertner.github.io/2020/10/use-gee-python-api-in-rstudio/
Previous blog posts are available within the blog archive. Feel free to connect or follow me on Twitter - @Mixed_Pixels.








