Intra-annual vegetation density changes of two public parks in Berlin, Germany
The blog post shows how the vegetation of two public parks changes seasonally. Multi-temporal satellite images were used to aggregate monthly histograms of vegetation index values. The visualization was inspired by this tweet from Christopher Doyle.
 Intra-annual vegetation density changes of two public parks in Berlin, Germany.
Intra-annual vegetation density changes of two public parks in Berlin, Germany.
Green spaces in the middle of the city
The topic in this blog post is Berlin, the pulsating metropolis everyone is talking about, at least that’s what most Berliner claim. But I won’t write about the club scene nor about hipster migration or the overcrowded, delayed local traffic.
Today, I write about two major parks in Berlins city center and show how their vegetation density changes seasonally over the course of the year. I am writing about the phenology.
Areas of Interest: Tiergarten and Tempelhofer Feld
Tiergarten is a popular inner-city park between the Kanzleramt and the Sony Business Center. About half of the Tiergarten consists of meadows the other half of woodland. The park is 210 ha in size and is among the largest urban gardens of Germany. Only Munich’s Englischer Garten and the Tempelhofer Feld, also in Berlin, are larger.
Now that I’m on it, the second park under investigation is the Tempelhofer Feld. The Tempelhofer Feld is with 355 ha the largest inner-city open space in the world. It is dominated by meadows but has at least 20 hectares of endangered and protected dry sand grassland and 27 hectares of plain oatmeal meadow.
Data generation in Earth Engine
The data basis for the illustration of the seasonal density change in the park vegetation were satellite images. Here, I used all USGS provided images (> 260) of Landsat 8 Collection 1 Tier 1, for each of the two parks, determined the Normalized Difference Vegetation Index (NDVI) and created a monthly NDVI histogram for all cloudless pixels depending on the date of recording and exported the result as .geojson. Very helpful was this page in the GEE Documentation.
In order to run the code you need access to the park polygons. Hereby I share the links to the Tiergarten und Tempelhof shapes, so everybody has read access to the asset and will be able to view und use them in the code editor.
GetData
// Javascript
// This function gets NDVI from a Landsat 8 image.
var addNDVI = function(image) {
return image.addBands(image.normalizedDifference(['B5', 'B4']));
};
// This function masks cloudy pixels.
var cloudMask = function(image) {
var clouds = ee.Algorithms.Landsat.simpleCloudScore(image).select(['cloud']);
return image.updateMask(clouds.lt(25));
};
// Load a Landsat collection, map the NDVI and cloud masking functions over it.
var collection = ee.ImageCollection('LANDSAT/LC08/C01/T1_TOA')
.filterBounds(Tiergarten.geometry())
.filter(ee.Filter.calendarRange(12,12, "month")) // only December
.map(addNDVI)
.map(cloudMask);
// Reduce the collection to the medan of each pixel and display.
var medianImage = collection.reduce(ee.Reducer.median());
// Reduce the region. The region parameter is the Feature geometry.
var median_Dictionary = medianImage.select(['nd_median']).reduceRegion({
reducer: ee.Reducer.histogram({minBucketWidth: 0.01}),
geometry: Tiergarten.geometry(),
scale: 30,
bestEffort: true
});
var FC = ee.Dictionary({median_Dictionary:median_Dictionary});
var features = ee.Feature(null, {FC: FC});
var export_FC = ee.FeatureCollection([features]);
Export.table.toDrive({
collection: export_FC,
folder: 'GEE',
description: 'Tiergarten_12',
fileFormat: 'GeoJSON'
});
// This function gets NDVI from a Landsat 8 image.
var addNDVI = function(image) {
return image.addBands(image.normalizedDifference(['B5', 'B4']));
};
// This function masks cloudy pixels.
var cloudMask = function(image) {
var clouds = ee.Algorithms.Landsat.simpleCloudScore(image).select(['cloud']);
return image.updateMask(clouds.lt(25));
};
// Load a Landsat collection, map the NDVI and cloud masking functions over it.
var collection = ee.ImageCollection('LANDSAT/LC08/C01/T1_TOA')
.filterBounds(Tiergarten.geometry())
.filter(ee.Filter.calendarRange(12,12, "month")) // only December
.map(addNDVI)
.map(cloudMask);
// Reduce the collection to the medan of each pixel and display.
var medianImage = collection.reduce(ee.Reducer.median());
// Reduce the region. The region parameter is the Feature geometry.
var median_Dictionary = medianImage.select(['nd_median']).reduceRegion({
reducer: ee.Reducer.histogram({minBucketWidth: 0.01}),
geometry: Tiergarten.geometry(),
scale: 30,
bestEffort: true
});
var FC = ee.Dictionary({median_Dictionary:median_Dictionary});
var features = ee.Feature(null, {FC: FC});
var export_FC = ee.FeatureCollection([features]);
Export.table.toDrive({
collection: export_FC,
folder: 'GEE',
description: 'Tiergarten_12',
fileFormat: 'GeoJSON'
});Data preprocessing with R
In the following section I present the steps you have to take to get from an Earth Engine produced .json file to a the animated GIF. The following code chunks are written in R.
Load all necessary libraries
### R
library(needs) # install.packages("needs")
needs(devtools, RJSONIO, ggplot2, tweenr, stringr, plyr, matrixStats, gganimate)Load the data from the ‘GEE’ folder into a data.frame
### R
file <- list.files("GEE")
dd <- data.frame()
for(i in 1:length(file)) {
d_json <- RJSONIO::fromJSON(paste("GEE/", file[[i]], sep=""))
hist <- d_json$features[[1]]$properties$FC$median_Dictionary$nd_median$histogram
bucketMeans <- d_json$features[[1]]$properties$FC$median_Dictionary$nd_median$bucketMeans
df <- data.frame(bucketMeans, hist, file[[i]])
dd <- rbind(dd, df)
}
colnames(dd) <- c("bucketMeans", "hist","i") # rename d.f columns
head(dd, n = 2)
## bucketMeans hist i
## 1 -0.06514288 2 Tempelhof_01.json
## 2 -0.05500000 0 Tempelhof_01.jsondd is the result of the histogram reducer which divided the entire range of image values into a series of intervals—and then counted how many values fall into each interval.
bucketMeans represents the bin (intervals) and hist the value count and i is the file name.
Split column i and create a location and month column
### R
a <- data.frame(str_split(dd$i, "_"))
a <- t(a)
a <- data.frame(a)
a$X2 <- str_replace(a$X2, ".json", "")
dd$location <- a$X1 # column location
dd$month <- as.numeric(a$X2) # column month
dd$i <- NULL # remove original "i" column
dd$hist <- round(dd$hist, 0)
colnames(dd) <- c("x", "y", "location", "month")
str(dd)
## 'data.frame': 1733 obs. of 4 variables:
## $ x : num -0.0651 -0.055 -0.0439 -0.035 -0.0299 ...
## $ y : num 2 0 4 0 2 1 2 2 15 66 ...
## $ location: Factor w/ 2 levels "Tempelhof","Tiergarten": 1 1 1 1 1 1 1 1 1 1 ...
## $ month : num 1 1 1 1 1 1 1 1 1 1 ...
summary(dd)
## x y location month
## Min. :-0.1706 Min. : 0.00 Tempelhof :815 Min. : 1.000
## 1st Qu.: 0.1450 1st Qu.: 3.00 Tiergarten:918 1st Qu.: 4.000
## Median : 0.3251 Median : 25.00 Median : 7.000
## Mean : 0.3274 Mean : 57.09 Mean : 6.563
## 3rd Qu.: 0.5059 3rd Qu.: 70.00 3rd Qu.: 9.000
## Max. : 0.8316 Max. :486.00 Max. :12.000Round bin values and create dummy a data.frame
To make the visualization easier we need for each month the same amout of bins, so I create a dummy data.frame with all possible value combination between x (bins)
and month, starting from -0.2 until 0.9.
### R
dd$x <- round_any(dd$x, accuracy = 0.01, f = ceiling)
# 12 (month) * 111 (NDVI value) = 1332 in total (ideal world row numbers)
combn_NDVI <- seq(-0.2,0.9, by = 0.01)
combn_month <- seq(1,12 , by = 1)
tempelhof <- tiergarten <- data.frame(month = rep(combn_month, each = length(combn_NDVI)),
x = rep(combn_NDVI, 12),
y = 0)
tiergarten$location <- "Tiergarten"
tempelhof$location <- "Tempelhof"
df <- rbind(tiergarten, tempelhof)
dim(df)
## [1] 2664 4Merge GEE data and the dummy data.frame
### R
dat <- merge(x = dd, y = df, by = c("location", "month", "x"),
all.y = T)
dat[is.na(dat)] <- 0 # convert NA's to 0
dat$y.y <- NULL # delete uncessesary columns
colnames(dat) <- c("location", "time" ,"x", "y")Smooth the curve
### R
dat_temp <- dat[dat$location == "Tempelhof", ]
dat_tier <- dat[dat$location == "Tiergarten", ]
do_it <- function(dataset) {
models <- dlply(dataset, .(time), function(df) loess(y ~ x, data = df, span = 0.1))
predict(models[[1]])
x <- ldply(models, predict)
library(reshape)
xx <- melt(x, id = c("time"))
xx <- xx[order(xx$time),]
return(xx)
}
xx <- do_it(dat_temp)
dat_temp$y_loess <- round(xx$value, 1)
dat_temp$y_loess[dat_temp$y_loess < 0] <- 0 # remove values beyond zero
####
xx <- do_it(dat_tier)
dat_tier$y_loess <- round(xx$value,1)
dat_tier$y_loess[dat_tier$y_loess < 0] <- 0
tail(dat_tier, n = 2)
## location time x y y_loess
## 2663 Tiergarten 12 0.89 0 0
## 2664 Tiergarten 12 0.90 0 0Add the ease and group columns for tweenr
### R
create_ease_and_group_for_tween <- function(dataset) {
dataset$ease <- "linear"
dataset <- dataset[order(dataset$x),]
dataset$group <- rep(1:length(combn_NDVI), each = 12)
dataset_tween <- tween_elements(data = dataset, time = 'time', group = 'group',
ease = 'ease', nframes = 111)
return(dataset_tween)
}
dat_temp_tween <- create_ease_and_group_for_tween(dat_temp)
dat_tier_tween <- create_ease_and_group_for_tween(dat_tier)
dat_tween <- rbind(dat_temp_tween, dat_tier_tween)
dat_tween$label <- round(dat_tween$time,0)
dat_tween$label <- month.name[dat_tween$label]
dim(dat_tween)
## [1] 24864 8
tail(dat_tween, n = 2)
## location time x y y_loess .frame .group label
## 109761 Tiergarten 12 0.77 0 0 111 98 December
## 110881 Tiergarten 12 0.78 0 0 111 99 DecemberCalculate a median for every point in time
### R
gg_df <- ddply(dat_tween, .(location, time), mutate,
weighted_median = weightedMedian(x,y_loess))
tail(gg_df, n = 2)
## location time x y y_loess .frame .group label
## 24863 Tiergarten 12 0.77 0 0 111 98 December
## 24864 Tiergarten 12 0.78 0 0 111 99 December
## weighted_median
## 24863 0.2403083
## 24864 0.2403083Plot the graph with ggplot + tweenr
Create the ggplot + tweenr histogram animation for the Tempelhofer Feld and the Tiergarten
### R
p <- ggplot(data = gg_df) +
# add label for time (month)
geom_text(data = gg_df[which(gg_df$location == 'Tiergarten'),],
aes(x = 0.42, y = 400, frame = .frame, label = label),
fontface = "bold", size = 20, color = "#d9d9d9") +
# add median point for Tempelhof
geom_segment(data = gg_df[which(gg_df$location == 'Tempelhof'),],
aes(x = weighted_median, y = 0, xend = weighted_median, yend = 490,
frame = .frame), size = 1, colour = "#737373") +
# add label for time (month)
geom_text(data = gg_df[which(gg_df$location == 'Tempelhof'),],
aes(x = weighted_median, y = 540, frame = .frame,
label = "Median\nTempelhofer Feld"),size = 5, colour = "#737373") +
# add median point for Tiergarten
geom_segment(data = gg_df[which(gg_df$location == 'Tiergarten'),],
aes(x = weighted_median, y = 0, xend = weighted_median, yend = 490,
frame = .frame), colour = "#737373", size = 1) +
# add median text for Tiergarten
geom_text(data = gg_df[which(gg_df$location == 'Tiergarten'),],
aes(x = weighted_median, y = 540, frame = .frame,
label = "Median\nTiergarten"),size = 5, colour = "#737373") +
# add NDVI ribbon for Tempelhof
geom_ribbon(data = gg_df[which(gg_df$location == 'Tempelhof'),],
aes(x = x, ymin = 0, ymax = y_loess, fill = "#2CA02C",frame = .frame),
alpha = 0.6) +
# add NDVI ribbon for Tiergarten
geom_ribbon(data = gg_df[which(gg_df$location == 'Tiergarten'),],
aes(x = x, ymin = 0, ymax = y_loess, frame = .frame, fill = "#FF7F0E"),
alpha = 0.6) +
labs(x = "", y = "") + theme_bw() + xlim(-0.1, 0.9) + ylim(0, 600) +
scale_fill_manual(values = c("#2CA02C", "#FF7F0E"),
labels = c("Tempelhofer Feld ", "Tiergarten")) +
scale_x_continuous(breaks = c(0, 0.25, 0.375, 0.5, 0.75),
labels = c("Low", "", "Medium","", "High")) +
theme(legend.position = "bottom",
legend.title = element_blank(),
axis.text.x = element_text(size = 15, colour ="#737373"),
axis.line.y = element_blank(),
axis.line.x = element_blank(),
panel.border = element_blank(),
axis.text.y = element_blank(),
axis.ticks = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
strip.background = element_blank(),
legend.text = element_text(size = 15, colour ="#737373", face = "bold"),
legend.key.size = unit(1.5, 'cm')) +
guides(fill = guide_legend(keywidth = 3.5, keyheight = 1.5))
animation::ani.options(interval = 1/5)
gganimate(p, 'Histogram_Tiergarten_Tempelhof_animated_0_01.gif',
title_frame = F,
ani.width = 800,
ani.height = 400)
## Scale for 'x' is already present. Adding another scale for 'x', which
## will replace the existing scale.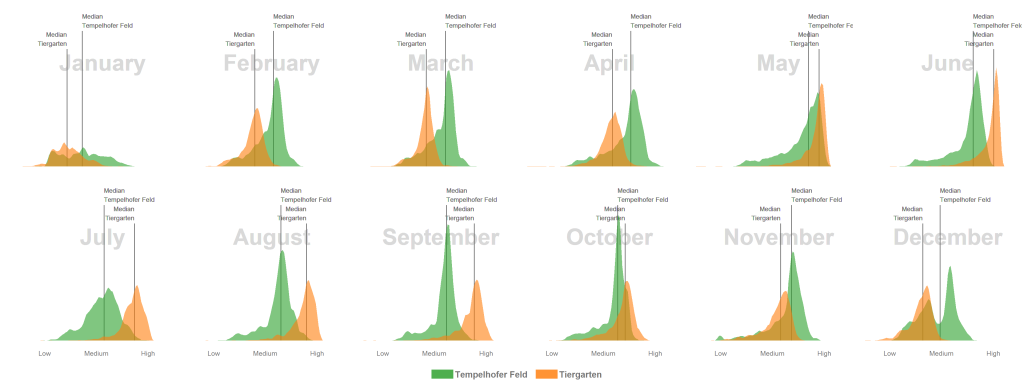 Intra-annual vegetation density changes of two public parks in Berlin, Germany
Intra-annual vegetation density changes of two public parks in Berlin, Germany
Intra-annual vegetation density changes
Unfortunately I don’t have any field data to validate my comments on the graphic here, so please use caution.
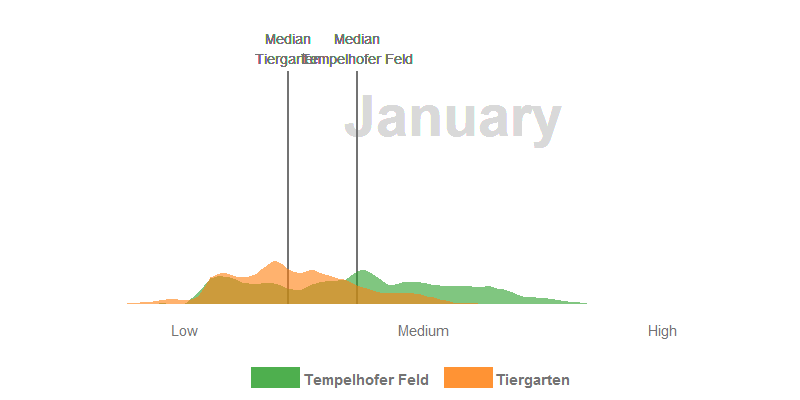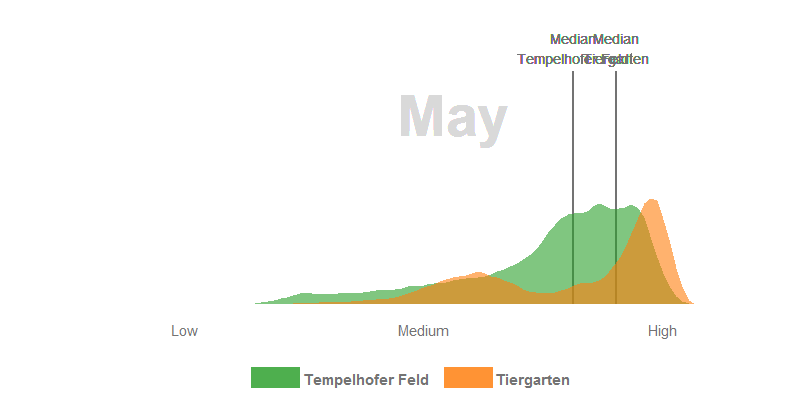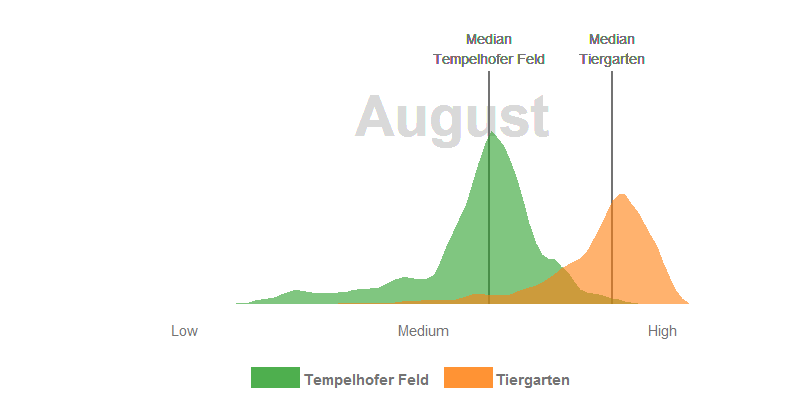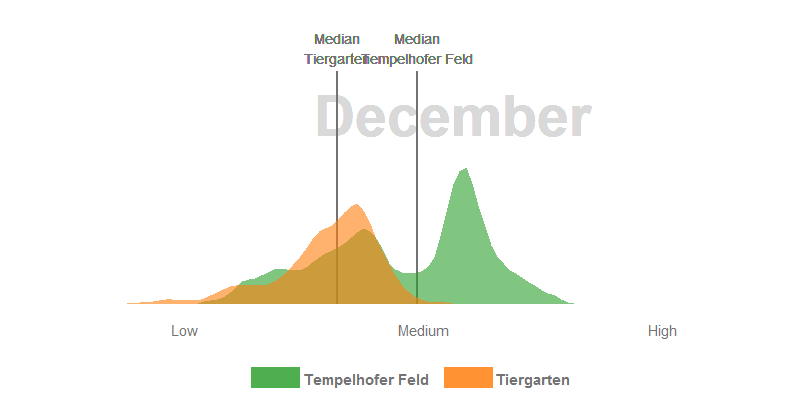
- The median NDVI values of the Tempelhofer Feld are higher in the winter and spring months than in the Tiergarten. The process of photosynthesis in the sprouts is reduced to a minimum. However, the sprouts of the grasses grow actively at temperatures > 10 °C and we’ve had a few days over the last few years far above these temperatures. Furthermore, the Tiergarten is stocked +/- 50% with deciduous trees and those lose their leaves in autumn. The autumn leaf also no longer forms a new chlorophyll and thus gradually loses its green colour, the NDVI decreases.
- In June, all the trees in the Tiergarten have turned out their leaves and the meadows grow superbly on the Tempelhofer Feld. Nevertheless, the multi-layered canopy roof in the Tiergarten usually has a higher leaf-area and canopy density which have generaly a positive relationship with the NDVI. In consequence, the Tiergarten has higher NDVI values in the summer months.
- In January there are significantly fewer values in the histogram than in summer. The reason for this is that the cloud pixels were filtered out during the histogram calculation.
Bottom line
You can cite this blogpost using: Gärtner, Philipp (2017/11/07) “Intra-annual vegetation density changes of two public parks in Berlin, Germany”, https://philippgaertner.github.io/2017/10/tempelhof_tiergarten/
If you have any questions, suggestions or spotted a mistake, please use the comment function at the bottom of this page.
Previous blog posts are available within the blog archive. Feel free to connect or follow me on Twitter - @Mixed_Pixels.
Data Source
- Park Polygons: The park polygons come from Michael Hörz berlin-shapes github repository.
- Satellite Data: The Landsat 8 Collection 1 Tier 1 calibrated top-of-atmosphere reflectance datat are provided by the USGS and made available by google as ImageCollection.

